Level: Basic
Required Knowledge: None
Keywords: Stream, Characterization, Library
Pipe-It Utilities: Streamz, Strexzel
This tutorial will focus on populating the Streamz Library of a Pipe-It project with an EOS characterization. Its usage together with a stream file will be described.
You can download the required input files here. You will need to login to the server using your existing username and password or register as new user. Download the file : "Manipulate_a_Stream_EOS_Characterization.zip" and un-zip it on your disk.
The folder contains a Pipe-It project called Tutorial_EOS_Characterization.ppv that we will use as a starting point. Open this Pipe-It project.
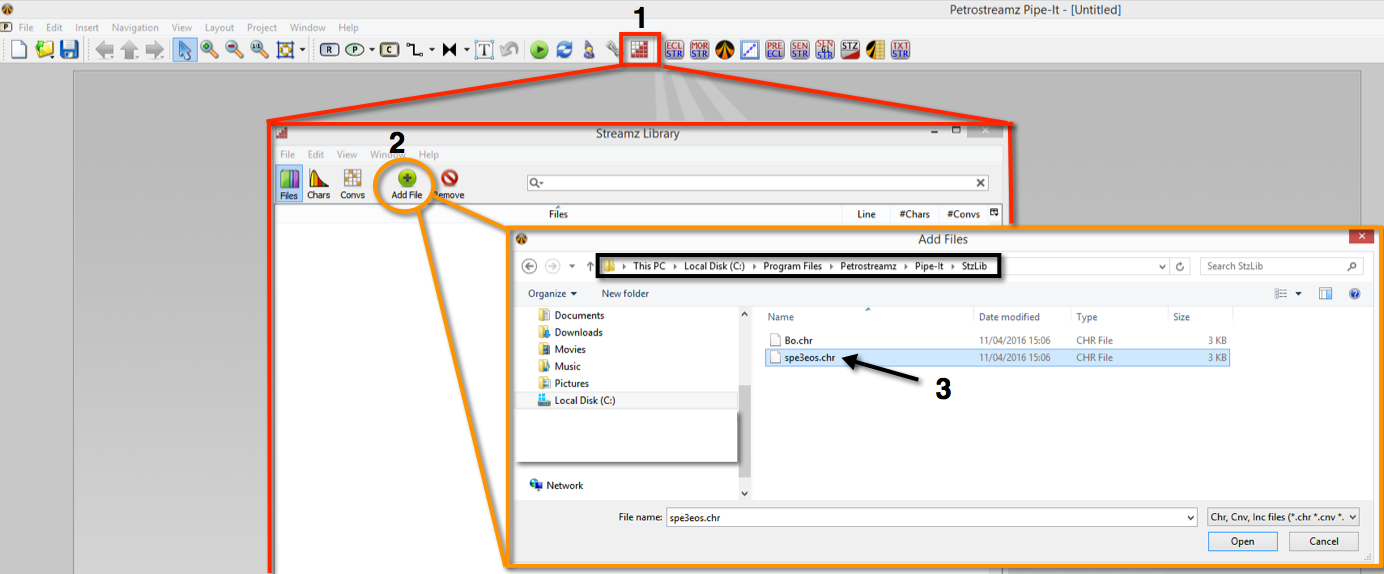
Open the Streamz Library by clicking on its icon in the task bar. It is empty at the moment. Choose to add a file. Instead of looking for one in the current folder let us use something provided by Pipe-It installation. Please navigate to the Pipe-It installation folder, typically c:\Program Files (x86)\Petrostreamz\Pipe-It\ and then to a folder called Stz Lib?. Just select the spe3eos.chr file. This file is scanned for import into the library and we can just say OK to continue.
This file is now imported in the Streamz Library. We can have a brief look at its content by double-clicking the entry. It opens in a text editor, e.g. Textpad. For the help of a learning Pipe-It user, this file has been documented by using comments (Asterix as the first character on the line). Users with knowledge of equation of state modeling will recognise entries like MW for Molecular Weight, PC for Critical Pressure, etc... In this example, a table of Binary Interaction Parameters (keyword "BINARY") is also included. With these properties defined, Pipe-it has the ability to perform efficient and accurate EOS. modeling such as phase separation. We see that this characterization comprises of 9 components.
A characterization is used in combination with a stream file containing same components as described by the characterization. A stream file is made available on the root folder of this tutorial project. It is called EOSstream.str. Locate it in your Explorer and drag it onto the canvas. Double-click on the file to open it in Strexzel. It has the same 9 components than the characterization. The molar quantities of each component are listed for each stream, which in this case corresponds to production for certain time steps from a simulator for a well named PROD. This file is extracted from the SPE paper 121252 called Model-Based Integration and Optimization – Gas Cycling Benchmark.
We need to tell Pipe-It that this stream file will use the characterization just imported into the library. To do so, right-click the resource and select "Set Characterization" (or just use the hot key F3). spe3EOS is the only characterization available. Note that if there were many characterizations in the Library, you could use the search bar to narrow down to the ones that you are looking for.
Now the EOSstream.str resource is ready for Streamz Process tasks in Pipe-It.
We can plot the stream file by right-clicking on the file and selecting "Plot Streams File...". The variation of moles per day of each component is plotted immediately with time. The plot represents the variation of the stream composition with time.
The Streamz Copier will be used to create a stream file containing molar fractions. Drop the Streamz Copier from the task bar on the canvas and connect it to the input stream file.
We need to create a new output stream resource to gather the molar fractions. One efficient way is to make a copy-paste of the input resource. To do this, right-click on the resource and select "Copy". Right-click again on the canvas and select "Paste".
A new resource shows on the canvas, we need now to associate it with a file on the disk. Right-click on the resource and select "Set Resource File..."The stream file doesn't exist yet. We'll call it EOSstream_fraction.str. Note that since we made a copy, the characterization is already set.
In order to generate molar fractions, we specify some options of the Copier by double-clicking on it. Select "Normalize" and close the Process. This option divides each component molar quantity by the sum of the molar quantities, giving the mole fraction.
The process is now all set to be ran. Click on the "Run" button on in the task bar. To compare the generated file with the input stream, you can double-click on it to look directly at the values calculated. You can also plot the file similarly to what was done previously. Side-by-side plots show quite different trends for molar quantities and mole fractions versus time. The plots below are focusing on C1.
Note: other manipulations like scaling, which can be used to implement unit conversions, or filtering can be applied in the Copier. In the Advanced tab, streams can be multiplied and/or divided by one or more variables that are accompanying the quantities of the streams. Such variables will typically be changing with each stream unlike a scaling with a scalar value.
The Single-Stage Separation that we will describe now gives the moles of each component in the gas phase and in the liquid phase after separation. There is one input stream file and two output stream files. Insert the Streamz Separator from the tool bar as shown below.
To create the resources, click on the resource icon on the tool bar and click again on the canvas. While the resource is selected, press the “F” hot-key to set the file name. Call them EOSstream_gas.str for the vapour and EOSstream_oil.str for the liquid. Connecting the Streamz Separator is similar to the Copier, although you need to connect the two output stream resources. Use the Connector icon to connect the Streamz Separator to the input and output resources.
Now we need to set the characterization. Since they will have the same characterization we can set the characterization for both resources simultaneously. Select both resources, right-click and say "Open Items Properties..." (hot-key is Ctrl-i). In the Items Properties window, select the Model tab and select the check-box “Apply to All” before selecting the appropriate characterization: SPE3EOS. Then select Apply and OK.
To set the conditions of the separator, open the process and make sure we are on the Separator page. By default, a new separator is setup to use the EOS Flash Type with a Standard Conditions of 1 atm pressure and 60°F temperature. Other methods of separation are possible: Split Factors and K-Values. Basic and Advanced options like the Streamz Copier are also available here. We'll keep the default conditions for this first calculation.
Launch the project (run icon on the tool bar). The moles of component that partitions into the gas and into the oil at the separator conditions are calculated. Double-click on the output oil and gas stream files to look at the data. Similarly to before, you can easily plot the results.
Using a series of processes it is possible to accurately and closely duplicate a full-fledged surface processing facility. Such an implementation is outside the scope of this tutorial.
One Streamz Processing that is possible without the need of conversions is to obtain the mass rates of the streams at any stage. This is accomplished by a simple Streamz Copier with an appropriate output resource connected to a file. Insert a Streamz Copier and connect the EOSstream_oil.str as the input stream. Create a new resource. Call it EOSstream_oil_mass.str. Set the characterization to SPE3EOS as we did it previously. Connect the new resource as the output stream of the Copier. Open the Copier and specify a conversion, but rather than the default Auto method, select Manual and then double-click the green box to set a self convertor. We choose to go from the Built-in Moles to Mass Convertor, which internally uses the Molecular Weights of the components in the characterization to perform the conversion.
We run the project again to get this resource up to date and then open it in Strexzel giving us the mass rates of each component.
We imported an EOS. characterization and attached it to a compositional stream file. We computed the mole fraction. We performed a single stage separation. Finally we converted the molar rates of product oil to mass rates.